
IMAGE: Lupinus angustifolius (a) and Arabidopsis thaliana (b). view more
Credit: Chiba University
Plants have evolved to produce a variety of compounds that vary from species to species. These compounds play a key role for the survival strategies of plants. Compounds that are bitter and poisonous protect the plants from being eaten by insects and animals. Meanwhile, compounds that have good smell or colorful pigments attract insects carrying pollens.
Among the compounds produced by plants, some substances have medicinal uses due to their physiological effects on the human body. Researchers have reported that matrine can be used as a depressant of the central nervous system, similar to morphine, and huperizine A can also be used as a supplement in the treatment of Alzheimer's disease. These compounds are called alkaloids, which are also the metabolites produced from amino acid lysine in certain species of plants.
Now, how has the system of lysine-derived alkaloids been evolved in plants? In 2016, the research team at Chiba University discovered that plants producing lysine-derived alkaloids have particular enzymes that possess bifunctional decarboxylating activity toward ornithine and lysine (lysine/ornithine decarboxylase). These enzymes have been evolved by a micro-mutation from the ancient enzyme (ornithine decarboxylase), which is essential for the production of polyamine used in fundamental and biological activities, such as cell division, in various organisms. These enzymes can decarboxylate and activate amino acid lysine and then send it into alkaloid production in plant cells. The micro-mutation is crucial for metabolism producing alkaloids and is common for plants using lysine as a material for alkaloid production.
Since this discovery in 2016, the research team has been expecting that the micro-mutated enzyme is a key factor in the plants' evolution for producing a certain type of alkaloid given that this mutation can be seen only in plant species producing lysine-derived alkaloid.
Finally, the research teams at Chiba University, RIKEN, and Kazusa DNA Research Institute confirmed in a paper published in the Plant Journal on July 31, 2019 that the alkaloids were generated by inserting the lysine/ornithine decarboxylating enzyme gene from Lupinus angustifolius producing lysine-derived alkaloid into Arabidopsis thaliana (A. thaliana). The team also identified the type of metabolites that were newly generated in A. thaliana, which is a model plant initially without the function to produce any alkaloid.
It is interesting to note that the research team recreated the metabolic evolution of the plant producing alkaloids in a model plant. A. thaliana was transformed to produce alkaloids by inserting the gene of the enzyme to switch the metabolic flow. The expression of the gene introduced in A. thaliana resulted in cadaverine (1,5-diaminopentane) production from lysine, which is further metabolized by endogenous enzymes. These processes resulted in the generation of new alkaloid related metabolites; 5-aminopentanal, 5-aminopentanoic acid, and δ-valerolactam by A. thaliana.
It is a noteworthy methodological development that the research team successfully identified the newly generated substances by detecting the changes in metabolites before and after introducing the gene, which is a challenging task given that plants usually contain several thousands of metabolites.
"We were able to change the metabolic flow by manipulating one of the genes in the plant, and also identify what kinds of alkaloids generated in the cells, which is a big step for the next discovery," said Mami Yamazaki, Associate Professor of Chiba University, who led the experiment. "Our research has opened a possible way to produce new compounds, which don't exist in nature yet, by performing similar genetic engineering on plants with different potentials of metabolic ability. Such expansion of chemical diversity is an important theme required for the development of the seeds in drug industries".
The research team is hoping that elucidating the metabolic mechanism in plants with the newly developed method will result in the stable supply of medical ingredients within the next several years.
###
REFERENCE
Yohei Shimizu, Amit Rai, Yuko Okawa, Hajime Tomatsu, Masaru Sato, Kota Kera, Hideyuki Suzuki, Kazuki Saito, and Mami Yamazaki, "Metabolic diversification of nitrogen-containing metabolites by expression of a heterologous lysine decarboxylase gene in Arabidopsis" The Plant Journal, July 31, 2019, doi: 10.1111/tpj.14454
WEBSITE
Research Lab for Phytochemical Plant Molecular Sciences, Chiba University http://www.
CONTACT
Saori Tanaka
Research Administrator for Communications
Institute for Global Prominent Research, Chiba University Tel: +81 (0)43 290 3022
Email: saori.tanaka@chiba-u.jp
Jens Wilkinson
RIKEN International Affairs Division Tel: +81-(0)48-462-1225
Email: pr@riken.jp
Public Relations and Research Promotion Group
Kazusa DNA Research Institute Tel: +81-438-52-3930
Fax: +81-438-52-3931
Email: kdri-kouhou@kazusa.or.jp


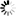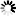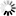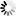

修改评论